Flow cytometry#
You’ll learn how to manage a growing number of flow cytometry data batches as a single queryable dataset.
Specifically, you will
read a single
.fcsfile as anAnnDataand seed a versioned dataset with it (, current page)
append a new data batch (a new
.fcsfile) to create a new version of the dataset ()
Setup#
!lamin init --storage ./test-facs --schema bionty
Show code cell output
✅ saved: User(id='DzTjkKse', handle='testuser1', email='testuser1@lamin.ai', name='Test User1', updated_at=2023-10-04 16:42:12)
✅ saved: Storage(id='dSnMNy0e', root='/home/runner/work/lamin-usecases/lamin-usecases/docs/test-facs', type='local', updated_at=2023-10-04 16:42:12, created_by_id='DzTjkKse')
💡 loaded instance: testuser1/test-facs
💡 did not register local instance on hub (if you want, call `lamin register`)
import lamindb as ln
import lnschema_bionty as lb
import readfcs
lb.settings.species = "human"
💡 loaded instance: testuser1/test-facs (lamindb 0.55.0)
hello
ln.track()
💡 notebook imports: lamindb==0.55.0 lnschema_bionty==0.31.2 pytometry==0.1.4 readfcs==1.1.7 scanpy==1.9.5
💡 Transform(id='OWuTtS4SAponz8', name='Flow cytometry', short_name='facs', version='0', type=notebook, updated_at=2023-10-04 16:42:16, created_by_id='DzTjkKse')
💡 Run(id='E3HTifOWblJJ2zFrUEug', run_at=2023-10-04 16:42:16, transform_id='OWuTtS4SAponz8', created_by_id='DzTjkKse')
hello
within hello
Ingest a first file#
Access  #
#
We start with a flow cytometry file from Alpert et al., Nat. Med. (2019).
Calling the following function downloads the file and pre-populates a few relevant registries:
ln.dev.datasets.file_fcs_alpert19(populate_registries=True)
hello
hello
hello
hello
PosixPath('Alpert19.fcs')
We use readfcs to read the raw fcs file into memory and create an AnnData object:
adata = readfcs.read("Alpert19.fcs")
adata
AnnData object with n_obs × n_vars = 166537 × 40
var: 'n', 'channel', 'marker', '$PnB', '$PnE', '$PnR'
uns: 'meta'
It has the following features:
adata.var.head(10)
| n | channel | marker | $PnB | $PnE | $PnR | |
|---|---|---|---|---|---|---|
| Time | 1 | Time | 32 | 0,0 | 2097152 | |
| Cell_length | 2 | Cell_length | 32 | 0,0 | 128 | |
| CD57 | 3 | (In113)Dd | CD57 | 32 | 0,0 | 8192 |
| Dead | 4 | (In115)Dd | Dead | 32 | 0,0 | 4096 |
| (Ba138)Dd | 5 | (Ba138)Dd | 32 | 0,0 | 4096 | |
| Bead | 6 | (Ce140)Dd | Bead | 32 | 0,0 | 16384 |
| CD19 | 7 | (Nd142)Dd | CD19 | 32 | 0,0 | 4096 |
| CD4 | 8 | (Nd143)Dd | CD4 | 32 | 0,0 | 4096 |
| CD8 | 9 | (Nd144)Dd | CD8 | 32 | 0,0 | 4096 |
| IgD | 10 | (Nd146)Dd | IgD | 32 | 0,0 | 8192 |
Transform: normalize  #
#
In this use case, we’d like to ingest & store curated data, and hence, we split signal and normalize using the pytometry package.
import pytometry as pm
First, we’ll split the signal from heigh and area metadata:
pm.pp.split_signal(adata, var_key="channel", data_type="cytof")
'area' is not in adata.var['signal_type']. Return all.
adata
AnnData object with n_obs × n_vars = 166537 × 40
var: 'n', 'channel', 'marker', '$PnB', '$PnE', '$PnR', 'signal_type'
uns: 'meta'
Normalize the dataset:
pm.tl.normalize_arcsinh(adata, cofactor=150)
Note
If the dataset was a flow dataset, you’ll also have to compensate the data, if possible. The metadata should contain a compensation matrix, which could then be run by the pytometry compensation function. In the case here, its a cyTOF dataset, which doesn’t (really) require compensation.
Validate: cell markers  #
#
First, we validate features in .var using CellMarker:
validated = lb.CellMarker.validate(adata.var.index)
hello
❗ 13 terms (32.50%) are not validated for name: Time, Cell_length, Dead, (Ba138)Dd, Bead, CD19, CD4, IgD, CD11b, CD14, CCR6, CCR7, PD-1
We see that many features aren’t validated because they’re not standardized.
Hence, let’s standardize feature names & validate again:
adata.var.index = lb.CellMarker.standardize(adata.var.index)
validated = lb.CellMarker.validate(adata.var.index)
hello
hello
hello
❗ 5 terms (12.50%) are not validated for name: Time, Cell_length, Dead, (Ba138)Dd, Bead
The remaining non-validated features don’t appear to be cell markers but rather metadata features.
Let’s move them into adata.obs:
adata.obs = adata[:, ~validated].to_df()
adata = adata[:, validated].copy()
Now we have a clean panel of 35 validated cell markers:
validated = lb.CellMarker.validate(adata.var.index)
assert all(validated) # all markers are validated
hello
Register: metadata  #
#
Next, let’s register the metadata features we moved to .obs.
For this, we create one feature record for each column in the .obs dataframe:
features = ln.Feature.from_df(adata.obs)
ln.save(features)
hello
hello
hello
hello
hello
We use the Experimental Factor Ontology through Bionty to create a “FACS” label:
lb.ExperimentalFactor.bionty().search("FACS").head(2) # search the public ontology
| ontology_id | definition | synonyms | parents | molecule | instrument | measurement | __ratio__ | |
|---|---|---|---|---|---|---|---|---|
| name | ||||||||
| fluorescence-activated cell sorting | EFO:0009108 | A Flow Cytometry Assay That Provides A Method ... | FACS|FAC sorting | [] | None | None | None | 100.0 |
| FACS-seq | EFO:0008735 | Fluorescence-Activated Cell Sorting And Deep S... | None | [EFO:0001457] | RNA assay | None | None | 90.0 |
We found one for “FACS”, let’s save it to our in-house registry:
# import the FACS record from the public ontology and save it to the registry
facs = lb.ExperimentalFactor.from_bionty(ontology_id="EFO:0009108")
facs.save()
hello
We don’t find one for “CyToF”, however, so, let’s create it without importing from a public ontology but label it as a child of “is_cytometry_assay”:
cytof = lb.ExperimentalFactor(name="CyTOF")
cytof.save()
is_cytometry_assay = lb.ExperimentalFactor(name="is_cytometry_assay")
is_cytometry_assay.save()
cytof.parents.add(is_cytometry_assay)
facs.parents.add(is_cytometry_assay)
is_cytometry_assay.view_parents(with_children=True)
hello
hello

Let us look at the content of the registry:
lb.ExperimentalFactor.filter().df()
| name | ontology_id | abbr | synonyms | description | molecule | instrument | measurement | bionty_source_id | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||
| lh5Cxy8w | fluorescence-activated cell sorting | EFO:0009108 | None | FACS|FAC sorting | A Flow Cytometry Assay That Provides A Method ... | None | None | None | KDDy | 2023-10-04 16:42:25 | DzTjkKse |
| U71A1UvQ | CyTOF | None | None | None | None | None | None | None | None | 2023-10-04 16:42:25 | DzTjkKse |
| BD7l3T5M | is_cytometry_assay | None | None | None | None | None | None | None | None | 2023-10-04 16:42:25 | DzTjkKse |
Register: data & annotate with metadata  #
#
modalities = ln.Modality.lookup()
features = ln.Feature.lookup()
experimental_factors = lb.ExperimentalFactor.lookup()
species = lb.Species.lookup()
hello
hello
hello
hello
file = ln.File.from_anndata(
adata, description="Alpert19", field=lb.CellMarker.name, modality=modalities.protein
)
... storing '$PnE' as categorical
... storing '$PnR' as categorical
hello
hello
hello
hello
hello
hello
hello
hello
hello
file.save()
Inspect the registered file#
Inspect features on a high level:
file.features
hello
hello
within hello
hello
within hello
Features:
var: FeatureSet(id='Jc7jskcvIuIvLTtkZ2kt', n=35, type='number', registry='bionty.CellMarker', hash='ldY9_GmptHLCcT7Nrpgo', updated_at=2023-10-04 16:42:26, modality_id='Q6B6XiyC', created_by_id='DzTjkKse')
'CD85j', 'CD11c', 'Igd', 'CD38', 'CD16', 'CD94', 'CD20', 'CD27', 'CD127', 'Ccr7', 'CD86', 'CD11B', 'CD45RA', 'DNA2', 'CD25', 'Cd4', 'CD24', 'CD3', 'TCRgd', 'PD1', ...
obs: FeatureSet(id='0FXtA0ATGyrSy7GrZ5oS', n=5, registry='core.Feature', hash='W4a3vMNiHC_k-_XfHV8F', updated_at=2023-10-04 16:42:26, modality_id='zBZgVlDk', created_by_id='DzTjkKse')
Dead (number)
(Ba138)Dd (number)
Bead (number)
Time (number)
Cell_length (number)
Inspect low-level features in .var:
file.features["var"].df().head()
hello
within hello
| name | synonyms | gene_symbol | ncbi_gene_id | uniprotkb_id | species_id | bionty_source_id | updated_at | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||
| lRZYuH929QDw | CD85j | None | None | None | uHJU | EBWO | 2023-10-04 16:42:20 | DzTjkKse | |
| L0WKZ3fufq0J | CD11c | ITGAX | 3687 | P20702 | uHJU | EBWO | 2023-10-04 16:42:20 | DzTjkKse | |
| 0evamYEdmaoY | Igd | None | None | None | uHJU | EBWO | 2023-10-04 16:42:20 | DzTjkKse | |
| CR7DAHxybgyi | CD38 | CD38 | 952 | B4E006 | uHJU | EBWO | 2023-10-04 16:42:20 | DzTjkKse | |
| bspnQ0igku6c | CD16 | FCGR3A | 2215 | O75015 | uHJU | EBWO | 2023-10-04 16:42:20 | DzTjkKse |
Use auto-complete for marker names in the var featureset:
markers = file.features["var"].lookup()
hello
within hello
markers.cd14
CellMarker(id='roEbL8zuLC5k', name='Cd14', synonyms='', gene_symbol='CD14', ncbi_gene_id='4695', uniprotkb_id='O43678', updated_at=2023-10-04 16:42:20, species_id='uHJU', bionty_source_id='EBWO', created_by_id='DzTjkKse')
In a plot, we can now easily also show gene symbol and Uniprot ID:
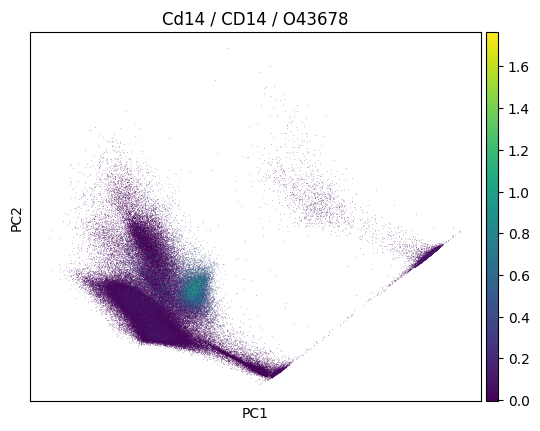
import scanpy as sc
sc.pp.pca(adata)
sc.pl.pca(
adata,
color=markers.cd14.name,
title=(
f"{markers.cd14.name} / {markers.cd14.gene_symbol} /"
f" {markers.cd14.uniprotkb_id}"
),
)
file.view_flow()
hello
within hello
hello
within hello
hello
within hello
hello
within hello
hello
hello
hello
hello
hello
hello
hello
within hello
hello
within hello
hello
within hello
hello
within hello
hello
hello
hello
hello
hello

Create a dataset from the file#
dataset = ln.Dataset(file, name="My versioned cytometry dataset", version="1")
dataset
hello
Dataset(id='dwdXtuUm7KR2XuBNSbsv', name='My versioned cytometry dataset', version='1', hash='VsTnnzHN63ovNESaJtlRUQ', transform_id='OWuTtS4SAponz8', run_id='E3HTifOWblJJ2zFrUEug', file_id='dwdXtuUm7KR2XuBNSbsv', created_by_id='DzTjkKse')
Let’s inspect the features measured in this dataset which were inherited from the file:
dataset.features
hello
hello
within hello
hello
within hello
Features:
var: FeatureSet(id='Jc7jskcvIuIvLTtkZ2kt', n=35, type='number', registry='bionty.CellMarker', hash='ldY9_GmptHLCcT7Nrpgo', updated_at=2023-10-04 16:42:26, modality_id='Q6B6XiyC', created_by_id='DzTjkKse')
'CD85j', 'CD11c', 'Igd', 'CD38', 'CD16', 'CD94', 'CD20', 'CD27', 'CD127', 'Ccr7', 'CD86', 'CD11B', 'CD45RA', 'DNA2', 'CD25', 'Cd4', 'CD24', 'CD3', 'TCRgd', 'PD1', ...
obs: FeatureSet(id='0FXtA0ATGyrSy7GrZ5oS', n=5, registry='core.Feature', hash='W4a3vMNiHC_k-_XfHV8F', updated_at=2023-10-04 16:42:26, modality_id='zBZgVlDk', created_by_id='DzTjkKse')
Dead (number)
(Ba138)Dd (number)
Bead (number)
Time (number)
Cell_length (number)
This looks all good, hence, let’s save it:
dataset.save()
Annotate by linking cytof & species labels:
dataset.labels.add(experimental_factors.cytof, features.assay)
dataset.labels.add(species.human, features.species)
hello
within hello
hello
within hello
hello
within hello
hello
within hello
hello
within hello
dataset.view_flow()
hello
within hello
hello
within hello
hello
hello
hello
hello
within hello
hello
within hello
hello
hello




